Imagine trying to read a library of books where every page contains billions of letters, yet the meaning behind those letters is hidden in complex patterns. This is what genomic research involves; it is a way of decoding the “book of life” written in DNA, RNA, and proteins.
NVIDIA, a global leader in AI and accelerated computing, is transforming this field by applying artificial intelligence (AI) techniques, including large language models (LLMs), to analyze and interpret biological data.
It has now gone even further; the latest Evo 2 by NVIDAI can now write genetic codes.
Evo 2: The Largest AI Model for Biology
Imagine a powerful tool that doesn’t just analyze the genetic code but can even write it. Evo 2 is the largest AI model ever created for biology. It is trained on 9.3 trillion DNA base pairs from over 128,000 genomes spanning all domains of life, including bacteria, archaea, eukaryotes, and bacteriophages.
Unlike traditional tools that focus on analyzing existing genomes, Evo 2 goes a step further by generating entire genomic sequences from scratch. This includes mitochondrial genomes, prokaryotic genomes, and even synthetic chromosomes for yeast and bacteria. It’s describing and designing biology.
Training Scale and Dataset
Evo 2 was trained on the largest dataset ever assembled for genomic modeling, including 9.3 trillion nucleotides (the building blocks of DNA and RNA). This dataset spans the entire tree of life, allowing Evo 2 to learn evolutionary patterns across species that humans have never observed before.
The training process involved NVIDIA’s DGX Cloud infrastructure, using over 2,048 H100 GPUs to process data at large scales. Compared to its predecessor, Evo 1, which focused mainly on prokaryotic genomes, Evo 2 incorporates eukaryotic genomes, which enables deeper insights into human biology and complex evolutionary relationships.
StripedHyena 2 Architecture
Evo 2 is built on StripedHyena 2, a novel deep learning architecture that combines convolutional filters with attention mechanisms. Unlike traditional Transformer models, which struggle with long DNA sequences, StripedHyena efficiently processes up to 1 million nucleotides in a single context window, eight times longer than Evo 1.
This allows Evo 2 to capture long-range interactions within DNA sequences, such as regulatory elements that control gene expression across distant regions.
Capabilities of Evo 2
1. Predicting Functional Effects of Mutations
Evo 2 excels at predicting how genetic mutations affect biological function.
For example:
- It achieved state-of-the-art performance in analyzing BRCA1 gene variants linked to breast cancer, accurately distinguishing harmful mutations from benign ones with over 90% accuracy.
- It predicts the impact of mutations in noncoding regions of DNA, which are areas traditionally considered “junk” but are crucial for regulating gene expression and disease development.
This capability could revolutionize precision medicine by identifying disease-causing mutations without the need for costly laboratory experiments.
2. Generative Abilities: Writing Genomes from Scratch
One of Evo 2’s most groundbreaking features is its ability to generate synthetic DNA sequences at the genome scale. It has successfully created:
- Synthetic yeast chromosomes.
- Mitochondrial genomes tailored for specific biological functions.
- Minimal bacterial genomes optimized for bioengineering applications.
Imagine designing programmable genomes that activate only in specific cell types, such as neurons or liver cells, to avoid side effects in gene therapies. Evo 2 makes this possible by precisely controlling genetic elements like promoters and enhancers.
3. Understanding Noncoding DNA
Noncoding DNA accounts for over 98% of the human genome and regulates critical processes like gene expression and chromatin accessibility.
Evo 2 autonomously learns features such as exon-intron boundaries and transcription factor binding sites without human guidance. This enables it to predict the functional impact of mutations in noncoding regions with unmatched accuracy.
4. Whole Genome Annotation
Evo 2 can annotate entire genomes, identifying key genetic elements such as start codons, splice sites, conserved regions, and transcription factor binding sites across diverse species and even those it hasn’t been explicitly trained on.
For example, Evo 2 successfully annotated the woolly mammoth genome from raw data without direct reference training.
Applications of Evo 2
- Synthetic Biology and Bioengineering
Evo 2 is paving the way for synthetic biology by enabling the computational design of new life forms. Its ability to generate programmable genomes could lead to innovations like designing synthetic organisms optimized for environmental cleanup or agricultural productivity and creating gene circuits capable of precisely controlling biological processes.
- Precision Medicine
Evo 2 could accelerate the development of personalized treatments for conditions like cancer, cardiovascular diseases, and rare genetic disorders by predicting disease-causing mutations and understanding noncoding DNA’s role in gene regulation.
- Evolutionary Biology
Evo 2 identifies evolutionary patterns across species by analyzing long-range dependencies within DNA sequences. This helps researchers uncover how genes evolve over millions of years and provides insights into biodiversity conservation efforts.
Why Evo 2 is a Game-Changer
- Open Source Accessibility: Evo 2 is fully open-source, including its model parameters, training code, inference code, and dataset (OpenGenome2). This democratizes access to powerful genomic tools, allowing researchers worldwide to innovate without barriers.
- Beyond Biology Analysis: Traditional AI models focus on analyzing biological data; Evo 2 goes beyond this by designing functional genomic systems from scratch. For example, it embedded Morse code into epigenomic designs as a proof-of-concept for programmable gene circuits, giving a perfect glimpse into future applications where biology becomes fully computational.
While Evo 2 was recently released, NVIDIA has been in the game for a long time. Let’s take a quick look back at some of NVIDIA’s breakthroughs in AI LLMs.
NVIDIA AtacWorks (2020)
NVIDIA’s AtacWorks leverages deep learning to enhance the analysis of ATAC-seq data, making it more accurate and efficient. By reducing noise and improving peak detection, AtacWorks enables researchers to extract meaningful insights from even the most challenging datasets.
Understanding Epigenomics
To understand epigenomics, imagine your DNA as a vast library of books. Each book represents a gene, and the words within these books are instructions for how your body functions.
However, not all books are open for reading at the same time. Epigenomics is like the librarian, deciding which books to open, which chapters to highlight, and which to keep closed based on what your body needs at any given moment.
Epigenomics studies changes in gene activity that do not alter the underlying DNA sequence but still affect gene expression.
Epigenomic changes are influenced by factors such as diet, exercise, aging, and environmental exposures. For instance, in diseases like cancer, abnormal DNA methylation patterns can silence tumor-suppressor genes or activate oncogenes, leading to uncontrolled cell growth.
Epigenomics is critical because it helps explain why identical twins with the same DNA can develop different diseases or why certain conditions run in families but skip generations.
It also has immense potential for personalized medicine, which can be archived through targeting specific epigenetic changes to treat diseases more effectively.
What is ATAC-seq?
ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) is a revolutionary method for mapping chromatin accessibility across the genome.
To understand its importance, consider chromatin a tightly packed bookshelf where only some books (genes) are accessible for reading (expression). ATAC-seq identifies which “books” are open and ready to be read by analyzing DNA regions accessible for regulatory proteins like transcription factors.
ATAC-seq is powerful but has limitations when working with small cell samples or low-quality data due to noise and low signal-to-noise ratios.
This is where NVIDIA’s AtacWorks comes in. It is a deep learning toolkit designed to enhance ATAC-seq data analysis by denoising noisy datasets and identifying regulatory peaks with high precision.
AtacWorks acts like an AI-powered microscope that sharpens blurry images of chromatin accessibility data, making it easier for researchers to identify meaningful patterns even from limited samples.
Key Features of AtacWorks
- Denoising Capability: AtacWorks uses convolutional neural networks (CNNs) to clean up noisy ATAC-seq data and produce high-resolution signals at single-base pair resolution.
- Peak Detection: It identifies genomic regions where chromatin is most accessible, which are key areas for understanding gene regulation.
- Efficiency: Traditional ATAC-seq analysis can take up to 15 hours on 32 CPUs; AtacWorks reduces this time to under 30 minutes using NVIDIA Tensor Core GPUs.
- Generalizability: Models trained with AtacWorks can analyze data from cell types not included in the training dataset, making it versatile across experiments.
BioNeMo Framework (2024)
NVIDIA’s BioNeMo uses AI to help scientists study DNA, proteins, and molecules more efficiently. It finds patterns and makes predictions that would take humans much longer to discover.
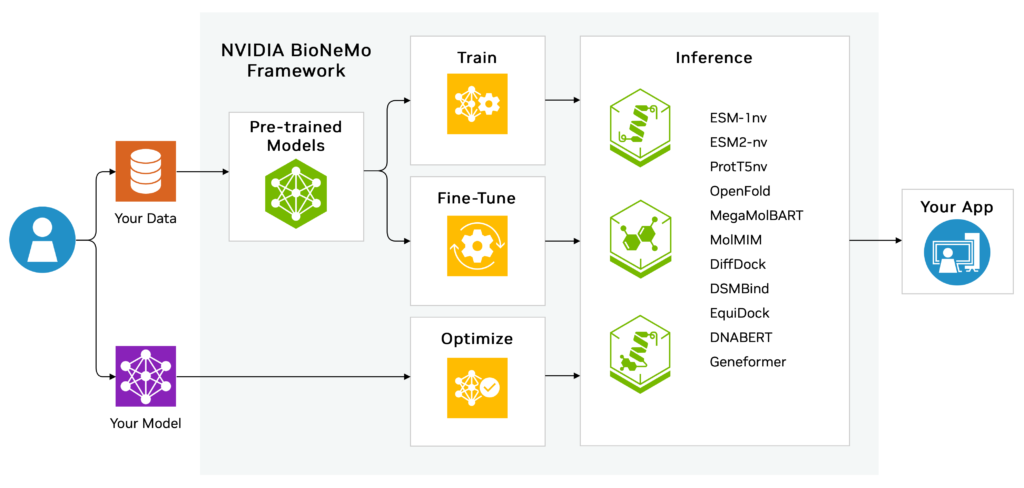
What is BioNeMo?
BioNeMo is NVIDIA’s framework for training and deploying large-scale language models tailored to biomolecular research. Just as AI models like ChatGPT understand human language, BioNeMo understands the “language” of biology, like DNA sequences, protein structures, RNA transcripts, and chemical compounds.
How Does BioNeMo Work?
BioNeMo builds on NVIDIA’s NeMo Megatron framework for GPU-accelerated training of large-scale models. It leverages transformer-based architectures capable of processing billions of parameters to uncover patterns in biomolecular data that would be impossible for humans to detect manually.
Capabilities of BioNeMo
BioNeMo supports tasks such as:
- Predicting protein structures using pre-trained models like AlphaFold2.
- Analyzing DNA sequences for disease-linked mutations or regulatory elements.
- Designing novel biomolecules optimized for therapeutic applications.
For instance, BioNeMo can generate new drug candidates by analyzing chemical structures represented in SMILES notation. It is a standardized way of describing molecules and predicting their properties, like solubility or binding affinity.
Traditional biomolecular research often involves small-scale models requiring extensive preprocessing steps. BioNeMo eliminates these bottlenecks by scaling up analysis capabilities while reducing computational costs through GPU acceleration. It enables researchers to work faster and more efficiently across drug discovery pipelines, from target identification to lead optimization.
BioNeMo LLM Service (2022)
NVIDIA’s BioNeMo LLM Service gives researchers an easy way to customize AI models for biomolecular studies using the cloud. It helps scientists fine-tune powerful models without needing advanced AI expertise or expensive computing resources.
What Does It Offer?
The BioNeMo LLM Service provides researchers with a cloud-based API for customizing generative AI models tailored to specific datasets and tasks. This flexibility allows scientists to adapt powerful pre-trained models without needing extensive computational resources or expertise in AI development.
Key Features
- Pretrained models can be fine-tuned using techniques like prompt learning or p-tuning, which require fewer examples than traditional methods.
- Supports tasks such as protein structure prediction and biomolecular property analysis.
Genomic LLMs: Performance and Applications
Genomic LLMs are specialized tools designed to understand the “grammar” of DNA, which are the rules governing how genes function and interact. By training these models on diverse datasets using supercomputers like Cambridge-1, NVIDIA has created genomic LLMs capable of tackling complex prediction tasks.
Applications
These models excel at tasks such as:
- Predicting enhancer/promoter sites, which are regions that regulate gene activity.
- Understanding transcription factor binding sites, which are key interactions in converting DNA into RNA.
Final Thought: The Biggest Discovery Today?
NVIDIA is undoubtedly onto something big that will transform humanity because, with these AI tools, researchers can analyze complex genomic datasets within hours instead of weeks. These generative AI models can also help design novel molecules optimized for therapeutic properties, drastically reducing costs and increasing success rates in clinical trials.
Looking ahead, NVIDIA plans to refine domain-specific LLMs further for applications like personalized medicine and advanced drug discovery. NVIDIA has partnered with industry leaders like AstraZeneca and the Broad Institute to push this further. Broader adoption by biotech startups, pharmaceutical companies, and academic researchers will likely drive innovation even faster.